Merge data
One other very common operation in R coding is to merge dataframes by common attributes (i.e., common column).
In the section about Data structure types, we sow how to merge matrices or dataframes using the rbind or cbind functions, which work when the different databases can be paired by simply appending one database to the other. For instance, when the columns are in exact order in the two databases.
Here we will use the merge function, or so call full_join from the dplr package, which works for cases when the data cannot be paired by simply appending one database to another. For instance, when the columns and/or rows are not in the exact order.
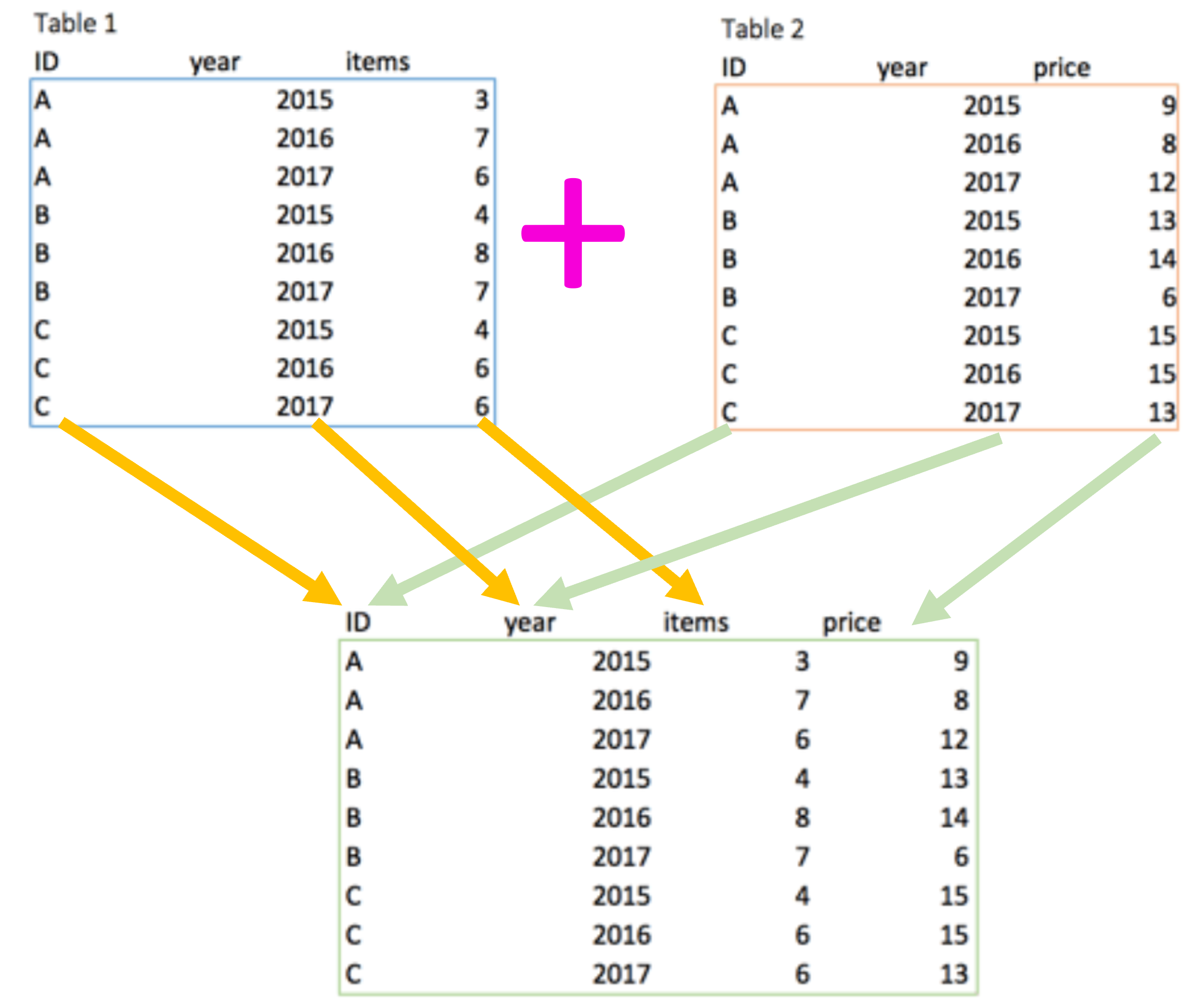
Figure 3.9: Merging function
Let’s use an example. As you know we have already loaded in this current section of R a database of the GPDs of the countries in the world.
What if I now collect another database about the average life expectancy per country, and want to put those two databases together?
Since the data comes from difference sources, chances are that they differ in the number of columns, in the order of the columns, in the number of rows, etc. Basically, you cannot use rbind or cbind in this case.
You could do that pairing of the two database by brutal force, copying the data from each country from one database, and pasting it in the same row for the given country in the other database. Say that operation takes you ten seconds for each country, and there are 250 countries, then that job will take you about 40 minutes, not to mention the chance for errors.
Oh, but remember that each country has 40 years of data. So multiplyy the 40 minutes times 40 years, and you end up copying and pasting for more than a day. Alternatively you have R do the merging for you. Lets try it.
First, lets load the data on the life expectancy, which I have placed in my Github folder
ExpectancyData=read.csv("https://raw.githubusercontent.com/Camilo-Mora/GEO380/main/Datasets/Countries_LifeExpectancy.csv")It is good practice to always check at least a portion of the database to check it has been loaded correctly,
## X country continent year lifeExp
## 1699 1699 Zimbabwe Africa 1982 60.363
## 1700 1700 Zimbabwe Africa 1987 62.351
## 1701 1701 Zimbabwe Africa 1992 60.377
## 1702 1702 Zimbabwe Africa 1997 46.809
## 1703 1703 Zimbabwe Africa 2002 39.989
## 1704 1704 Zimbabwe Africa 2007 43.487Lets check the GDP database as well.
## country continent year gdpPercap
## 1699 Zimbabwe Africa 1982 788.8550
## 1700 Zimbabwe Africa 1987 706.1573
## 1701 Zimbabwe Africa 1992 693.4208
## 1702 Zimbabwe Africa 1997 792.4500
## 1703 Zimbabwe Africa 2002 672.0386
## 1704 Zimbabwe Africa 2007 469.7093Ok, we now have two databases loaded in R, one for the data on GDP and the other for the data on life expectancy. Lets merge them together, like this:
Now lets try to read, that line of code, like this:

Figure 3.10: Merging function
If I translate this…basically, create a new dataframe called MergeData, in which I want to merge in full the database on GDPData and ExpectancyData by their common attribute country and year.
## country continent.x year gdpPercap X continent.y lifeExp
## 1699 Zimbabwe Africa 1982 788.8550 1699 Africa 60.363
## 1700 Zimbabwe Africa 1987 706.1573 1700 Africa 62.351
## 1701 Zimbabwe Africa 1992 693.4208 1701 Africa 60.377
## 1702 Zimbabwe Africa 1997 792.4500 1702 Africa 46.809
## 1703 Zimbabwe Africa 2002 672.0386 1703 Africa 39.989
## 1704 Zimbabwe Africa 2007 469.7093 1704 Africa 43.487